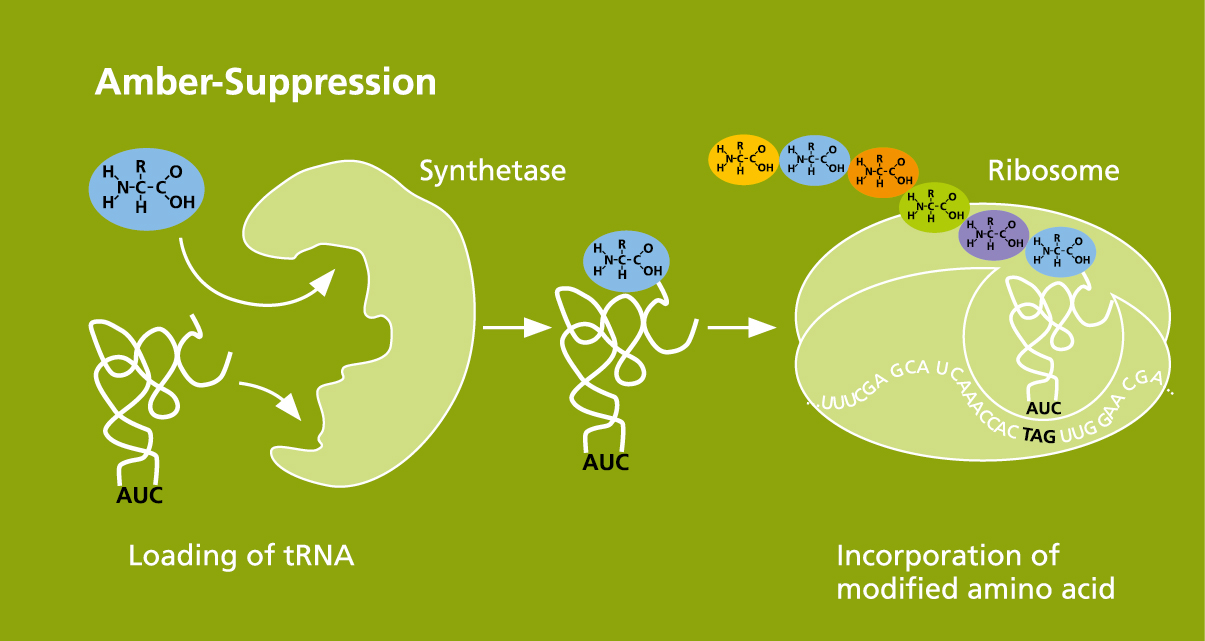
There are 20 basic amino acids that make up every protein in the natural world – with a few exceptions. Due to this limited number of amino acids, there are also limitations in terms of the biochemical and biophysical properties of the desired functional proteins.

The Cell-free Protein Synthesis Unit offers a reliable method for introducing chemically modified amino acids with highly diverse reactive groups into cell-free synthesized proteins at specific positions. Amber suppression is the underlying method for this. In addition, we use click chemistry to couple fluorescent dyes, sugar structures, PEGylations and biotinylations to the reactive groups of the amino acids being introduced. Our methods are particularly well-suited for structural analysis and functional identification of membrane proteins, as well as for the screening of new kinds of ligands and therapeutic substances. Proteins can also be selectively modified in this way to identify new functions that may be used for treatment purposes.
 Fraunhofer Institute for Cell Therapy and Immunology, Branch Bioanalytics and Bioprocesses IZI-BB
Fraunhofer Institute for Cell Therapy and Immunology, Branch Bioanalytics and Bioprocesses IZI-BB